[1]:
import os,sys
%matplotlib inline
import matplotlib.pylab as plt
import pickle
import numpy as np
plt.rcParams['figure.dpi'] = 100
plt.rcParams['savefig.dpi']=300
# sys.path.append(os.path.expanduser("~/Projects/Github/PyComplexHeatmap"))
from PyComplexHeatmap import *
use_pch_style() # or plt.style.use('default') to restore default style
# plt.rcParams
# import matplotlib; print(matplotlib.__version__)
[2]:
#set font to Arial using the following code
plt.rcParams['font.family']='sans serif'
plt.rcParams['font.sans-serif']='Arial'
# set pdf.fonttype to 42
plt.rcParams['pdf.fonttype']=42
Generate dataset¶
[3]:
#Generate example dataset (random)
df = pd.DataFrame(['GroupA'] * 5 + ['GroupB'] * 5, columns=['AB'])
df['CD'] = ['C'] * 3 + ['D'] * 3 + ['G'] * 4
df['EF'] = ['E'] * 6 + ['F'] * 2 + ['H'] * 2
df['F'] = np.random.normal(0, 1, 10)
df.index = ['sample' + str(i) for i in range(1, df.shape[0] + 1)]
df_box = pd.DataFrame(np.random.randn(10, 4), columns=['Gene' + str(i) for i in range(1, 5)])
df_box.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar = pd.DataFrame(np.random.uniform(0, 10, (10, 2)), columns=['TMB1', 'TMB2'])
df_bar.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_scatter = pd.DataFrame(np.random.uniform(0, 10, 10), columns=['Scatter'])
df_scatter.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_heatmap = pd.DataFrame(np.random.randn(30, 10), columns=['sample' + str(i) for i in range(1, 11)])
df_heatmap.index = ["Fea" + str(i) for i in range(1, df_heatmap.shape[0] + 1)]
df_heatmap.iloc[1, 2] = np.nan
[4]:
# add a missing value to sample4
df_heatmap.loc['Fea4','sample4']=np.nan
df_box.loc['sample4','Gene4']=np.nan
df_box
[4]:
| Gene1 | Gene2 | Gene3 | Gene4 | |
|---|---|---|---|---|
| sample1 | 0.786715 | -0.268172 | -0.165060 | -0.003303 |
| sample2 | 0.135651 | -0.108862 | -0.452932 | 0.542925 |
| sample3 | -0.569915 | -0.291013 | -0.551063 | 0.126623 |
| sample4 | -0.420822 | 0.200885 | 0.807041 | NaN |
| sample5 | 1.638377 | 1.261568 | -0.762235 | 2.018141 |
| sample6 | 0.074054 | -2.008517 | 1.742215 | -0.403439 |
| sample7 | -0.436391 | 0.810032 | -0.354438 | 0.122689 |
| sample8 | 1.561714 | -0.338679 | -1.749112 | -2.315677 |
| sample9 | 0.448826 | 0.220147 | -0.449316 | -1.141088 |
| sample10 | -0.353502 | 0.084744 | 0.912792 | 0.469271 |
Add selected rows labels¶
[72]:
#Annotate the rows with average > 0.3
df_rows = df_heatmap.apply(lambda x:x.name if x.sample4 > 0.5 else None,axis=1)
df_rows=df_rows.to_frame(name='Selected')
df_rows['XY']=df_rows.index.to_series().apply(lambda x:'A' if int(x.replace('Fea',''))>=15 else 'B')
row_ha = HeatmapAnnotation(
Scatter=anno_scatterplot(df_heatmap.sample4.apply(lambda x:round(x,2)),
height=12,cmap='jet',legend=False,grid=True,
legend_kws=dict(color='red')),
Line=anno_lineplot(df_heatmap.sample4.apply(lambda x:round(x,2)),
height=12,colors='red',linewidth=2,legend=False),
Bar=anno_barplot(df_heatmap.sample4.apply(lambda x:round(x,2)),
height=15,cmap='rainbow',legend=False),
selected=anno_label(df_rows,colors='red',relpos=(-0.05,0.4)),
label_kws={'rotation':30,'horizontalalignment':'left','verticalalignment':'bottom'},
axis=0,verbose=0)
col_ha = HeatmapAnnotation(
label=anno_label(df.AB, merge=True,rotation=10,
arrowprops = dict(visible=False),
),
AB=anno_simple(df.AB,add_text=True),
axis=1,
CD=anno_simple(df.CD,add_text=True),
EF=anno_simple(df.EF,add_text=True,
legend_kws={'frameon':True}),
G=anno_boxplot(df_box, cmap='jet',legend=False,grid=True),
verbose=0)
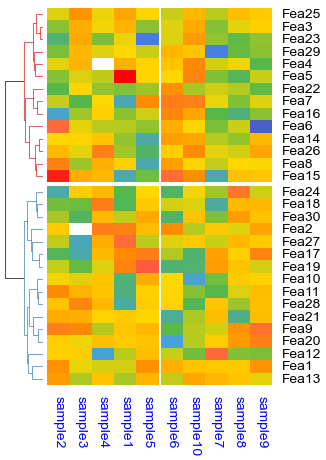
print(np.nanmin(df_heatmap),np.nanmax(df_heatmap))
plt.figure(figsize=(5.5, 6.5))
cm = ClusterMapPlotter(
data=df_heatmap, top_annotation=col_ha,right_annotation=row_ha,
col_cluster=True,row_cluster=True,
col_split=df.AB,row_split=2, z_score=0,vmin=-2.2,vmax=2.3,
col_split_gap=0.5,row_split_gap=0.8,
row_dendrogram=True,col_dendrogram=False,row_dendrogram_size=15,
show_rownames=False,show_colnames=True,
tree_kws={'row_cmap': 'Set1'},verbose=0,legend_gap=5,
cmap='RdYlBu_r',bezier=True,dotsize=2,
legend_kws=dict(label='test'), #label='values',
xticklabels_kws=dict(labelrotation=90,labelcolor='blue',labelsize=14,grid_color='red',bottom=True))
# for ax in cm.top_annotation.axes[-1,:]:
# ax.cla()
plt.savefig("example0.pdf", bbox_inches='tight')
plt.show()
print(cm.kwargs['vmin'],cm.kwargs['vmax'],cm.legend_kws)
-3.3541065464638127 2.5905806804216205
-2.2 2.3 {'label': 'test', 'vmin': -2.2, 'vmax': 2.3, 'center': None}
[6]:
cm.heatmap_axes
[6]:
array([[<Axes: >, <Axes: >],
[<Axes: >, <Axes: >]], dtype=object)
[7]:
cm.ax_heatmap.xaxis.get_tick_params()
[7]:
{'rotation': 90,
'left': True,
'right': False,
'labelleft': False,
'labelright': False,
'gridOn': False,
'labelcolor': 'blue',
'labelsize': 14,
'grid_color': 'red'}
Add annotations on the top of heatmap cells¶
[8]:
#Annotate the rows with average > 0.3
df_rows = df_heatmap.apply(lambda x:x.name if x.sample4 > 0.5 else None,axis=1)
df_rows=df_rows.to_frame(name='Selected')
df_rows['XY']=df_rows.index.to_series().apply(lambda x:'A' if int(x.replace('Fea',''))>=15 else 'B')
row_ha = HeatmapAnnotation(
S4=anno_simple(df_heatmap.sample4.apply(lambda x:round(x,2) if not pd.isna(x) else ''),
add_text=True,height=10,legend=False,
text_kws={'rotation':0,'fontsize':10,'color':'blue'}),
# Scatter=anno_scatterplot(df_heatmap.sample4.apply(lambda x:round(x,2)),
# height=10),
Test=anno_barplot(df_heatmap.sample4.apply(lambda x:round(x,2)),
height=18,cmap='rainbow',grid=True),
selected=anno_label(df_rows,colors='red'),
axis=0,verbose=0,#wgap=4,
label_kws={'rotation':0,'horizontalalignment':'left',
'verticalalignment':'bottom'})
col_ha = HeatmapAnnotation(
label=anno_label(df.AB, merge=True,rotation=15),
AB=anno_simple(df.AB,add_text=True),axis=1,
CD=anno_simple(df.CD,add_text=True,text_kws=dict(bbox={'boxstyle':'Circle','edgecolor':'white','fill':False},fontsize=8),
height=4.5),
EF=anno_simple(df.EF,add_text=True,
legend_kws={'frameon':False}),
Exp=anno_boxplot(df_box, cmap='turbo',grid=True),
verbose=0,#hgap=2
) #verbose=0 will turn off the log.
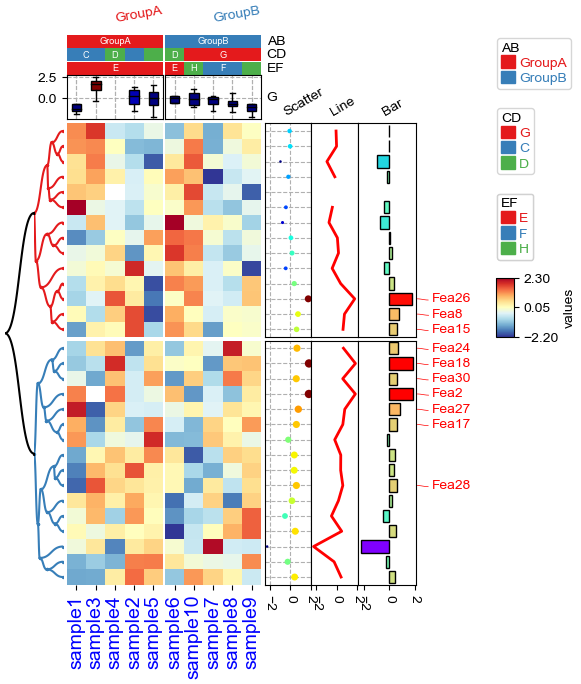
print(df.head())
print(df_box.mean(axis=1))
print(df_heatmap.head())
plt.figure(figsize=(6, 8))
cm = ClusterMapPlotter(
data=df_heatmap, top_annotation=col_ha,right_annotation=row_ha,
col_split=df.AB,
row_split=df_rows.XY,
#col_split_gap=3.5,row_split_gap=2.5,
col_cluster=True,row_cluster=True,
label='values',row_dendrogram=False,
show_rownames=True,show_colnames=True,
verbose=0,legend_gap=5,#legend_hpad=10,legend_vpad=5,
annot=False,fmt='.1g',linewidths=0.05,linecolor='gold',cmap='RdYlBu_r',
xticklabels_kws={'labelrotation':-45,'labelcolor':'blue'},
yticklabels_kws=dict(labelcolor='red'),#subplot_gap=8
)
#subplot_gap controls the gap between main heatmap and column or row annotations
plt.show()
print(cm.row_order)
print(cm.col_order)
AB CD EF F
sample1 GroupA C E 0.362690
sample2 GroupA C E -0.982457
sample3 GroupA C E 1.067007
sample4 GroupA D E -1.318330
sample5 GroupA D E 0.046514
sample1 -0.937345
sample2 0.047537
sample3 1.359393
sample4 -0.477066
sample5 -0.132495
sample6 -0.371260
sample7 -0.368397
sample8 -0.602725
sample9 -1.261328
sample10 -0.043332
dtype: float64
sample1 sample2 sample3 sample4 sample5 sample6 sample7 \
Fea1 -0.256096 1.294337 -0.085543 -0.243972 1.303379 0.755107 0.361045
Fea2 1.735609 0.316170 NaN 1.830172 0.544176 1.372135 0.351998
Fea3 0.733865 -0.798262 0.775947 0.002780 -0.826095 -0.162563 -0.860632
Fea4 0.781866 -0.127187 0.703247 NaN 0.164684 0.433726 -0.256959
Fea5 3.356756 -0.846319 -0.207551 -0.409454 0.172251 0.093536 -0.893011
sample8 sample9 sample10
Fea1 0.330225 1.236336 0.429788
Fea2 -0.153091 0.981548 -0.476542
Fea3 -0.143750 0.184459 0.851115
Fea4 -0.030188 -1.230343 1.477730
Fea5 -1.336817 -0.312624 1.645297
[['Fea24', 'Fea18', 'Fea30', 'Fea27', 'Fea20', 'Fea17', 'Fea19', 'Fea21', 'Fea28', 'Fea15', 'Fea26', 'Fea23', 'Fea25', 'Fea22', 'Fea16', 'Fea29'], ['Fea6', 'Fea8', 'Fea14', 'Fea2', 'Fea4', 'Fea3', 'Fea5', 'Fea12', 'Fea9', 'Fea10', 'Fea11', 'Fea13', 'Fea1', 'Fea7']]
[['sample2', 'sample3', 'sample4', 'sample1', 'sample5'], ['sample6', 'sample10', 'sample7', 'sample8', 'sample9']]
Only plot the annotations¶
[9]:
df = pd.DataFrame(['AAAA1'] * 5 + ['BBBBB2'] * 5, columns=['AB'])
df['CD'] = ['C'] * 3 + ['D'] * 3 + ['G'] * 4
df['F'] = np.random.normal(0, 1, 10)
df.index = ['sample' + str(i) for i in range(1, df.shape[0] + 1)]
df_box = pd.DataFrame(np.random.randn(10, 4), columns=['Gene' + str(i) for i in range(1, 5)])
df_box.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar = pd.DataFrame(np.random.uniform(0, 10, (10, 2)), columns=['TMB1', 'TMB2'])
df_bar.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_scatter = pd.DataFrame(np.random.uniform(0, 10, 10), columns=['Scatter'])
df_scatter.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar1 = pd.DataFrame(np.random.uniform(0, 10, (10, 2)), columns=['T1-A', 'T1-B'])
df_bar1.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar2 = pd.DataFrame(np.random.uniform(0, 10, (10, 2)), columns=['T2-A', 'T2-B'])
df_bar2.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar3 = pd.DataFrame(np.random.uniform(0, 10, (10, 2)), columns=['T3-A', 'T3-B'])
df_bar3.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar3.iloc[5,0]=np.nan
df_bar4 = pd.DataFrame(np.random.uniform(0, 10, (10, 1)), columns=['T4'])
df_bar4.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar4.iloc[7,0]=np.nan
print(df)
print(df_box.head())
print(df_scatter)
print(df_bar)
print(df_bar1)
print(df_bar2)
print(df_bar3)
print(df_bar4)
AB CD F
sample1 AAAA1 C -1.395423
sample2 AAAA1 C 0.315828
sample3 AAAA1 C -1.894856
sample4 AAAA1 D -0.207946
sample5 AAAA1 D -0.142276
sample6 BBBBB2 D 0.552056
sample7 BBBBB2 G -0.176148
sample8 BBBBB2 G 0.169970
sample9 BBBBB2 G -1.398254
sample10 BBBBB2 G -1.422476
Gene1 Gene2 Gene3 Gene4
sample1 0.170749 0.369818 -0.406722 0.831243
sample2 1.356510 -1.586601 0.360872 -1.808163
sample3 -0.142937 -0.402044 -1.372085 -0.006017
sample4 -1.960390 -2.046558 -0.034469 1.413408
sample5 0.462673 0.151571 0.420617 0.403127
Scatter
sample1 2.845265
sample2 3.092774
sample3 9.248764
sample4 5.926675
sample5 3.709297
sample6 3.385716
sample7 6.177794
sample8 2.923336
sample9 4.870179
sample10 1.979889
TMB1 TMB2
sample1 8.773112 1.653713
sample2 7.586741 7.328379
sample3 5.791634 4.394594
sample4 2.631353 2.556334
sample5 2.043007 6.890823
sample6 6.906687 7.230015
sample7 0.072210 0.240389
sample8 4.331631 8.055023
sample9 6.405677 8.152781
sample10 7.203812 3.805164
T1-A T1-B
sample1 6.962736 7.163614
sample2 0.599616 2.111658
sample3 1.991862 7.737146
sample4 5.307607 2.107476
sample5 5.391606 5.642582
sample6 9.262886 0.798720
sample7 0.149710 9.832642
sample8 5.501970 0.465060
sample9 4.053081 3.171087
sample10 5.727097 6.287463
T2-A T2-B
sample1 9.541359 0.763995
sample2 0.407472 5.127191
sample3 3.910844 7.810868
sample4 9.638641 4.979790
sample5 6.229322 4.687453
sample6 4.682800 2.185468
sample7 0.316455 2.642579
sample8 3.042201 0.237428
sample9 7.226572 4.233363
sample10 6.226147 5.776414
T3-A T3-B
sample1 6.367489 1.073648
sample2 8.109799 7.400489
sample3 0.510005 8.161129
sample4 1.897186 5.820624
sample5 4.127147 9.799400
sample6 NaN 5.438595
sample7 2.478326 1.461691
sample8 8.589167 1.485746
sample9 8.187439 0.822651
sample10 1.070760 6.860972
T4
sample1 5.839575
sample2 7.886911
sample3 8.504942
sample4 5.119021
sample5 1.859604
sample6 6.721850
sample7 0.545908
sample8 NaN
sample9 7.443164
sample10 1.677521
[10]:
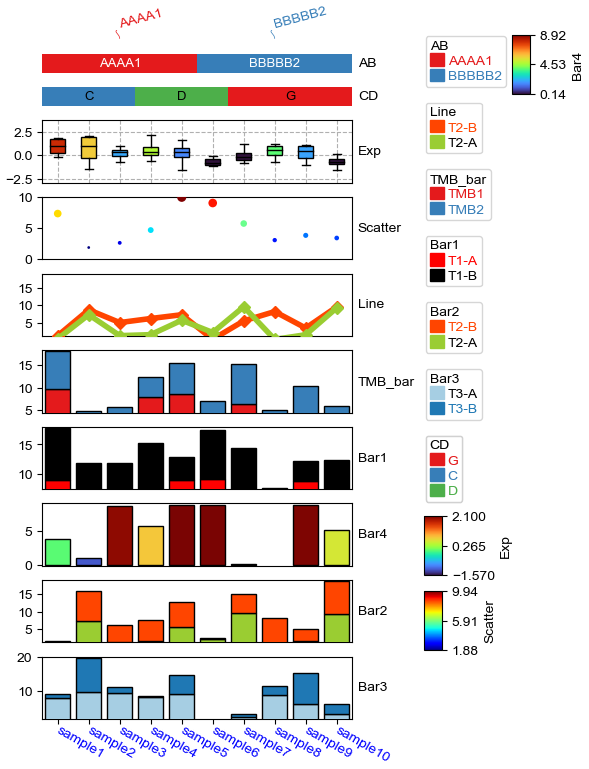
plt.figure(figsize=(4, 8))
col_ha = HeatmapAnnotation(
label=anno_label(df.AB, merge=True,rotation=15),
AB=anno_simple(df.AB,add_text=True), axis=1,
CD=anno_simple(df.CD, add_text=True,text_kws={'color':'black'}),
Exp=anno_boxplot(df_box, cmap='turbo',grid=True),
Scatter=anno_scatterplot(df_scatter,legend=True,grid=True),
Line=anno_lineplot(df_bar2,linewidth=4,colors={'T2-B':'orangered','T2-A':'yellowgreen'},
marker='D',legend=True), #colors=['orangered','yellowgreen']
TMB_bar=anno_barplot(df_bar,legend=True,cmap='Set1'),
Bar1=anno_barplot(df_bar1,legend=True,colors=['red','black']), #colors can be str, list, tuple or dict
Bar4=anno_barplot(df_bar4,legend=True,cmap='turbo'),
Bar2=anno_barplot(df_bar2,legend=True,colors={'T2-B':'orangered','T2-A':'yellowgreen'}),
Bar3=anno_barplot(df_bar3,legend=True,cmap='Paired'),
plot=True,legend=True,legend_gap=5,hgap=4,
# legend_order=False,
# legend_order=['AB','CD','Line','Bar1'],legend_width=20
)
col_ha.show_ticklabels(df.index.tolist(),fontdict={'color':'blue'},rotation=-30)
plt.show()
Starting plotting HeatmapAnnotations
Collecting annotation legends..
Change orentation to down and add extra space¶
[11]:
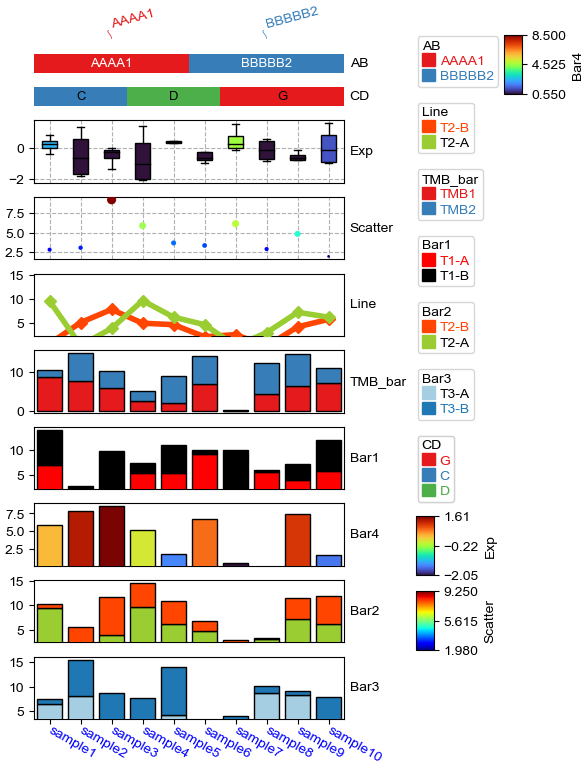
plt.figure(figsize=(4, 8))
row_ha = HeatmapAnnotation(
TMB_bar=anno_barplot(df_bar,legend=True,cmap='Set1'),
Bar1=anno_barplot(df_bar1,legend=True,cmap='Dark2'),
Bar4=anno_barplot(df_bar4,legend=True,cmap='turbo'),
Bar2=anno_barplot(df_bar2,legend=True,cmap='tab10'),
Bar3=anno_barplot(df_bar3,legend=True,cmap='Paired'),
Scatter=anno_scatterplot(df_scatter),
Exp=anno_boxplot(df_box, cmap='turbo',legend=True,grid=True),
CD=anno_simple(df.CD, colors={'C': 'red', 'D': 'gray', 'G': 'yellow'},
add_text=True,legend=True,text_kws={'color':'black'}),
AB=anno_simple(df.AB,add_text=True,legend=True),
label=anno_label(df.AB, merge=True,rotation=-15),
plot=True,plot_legend=False,legend_hpad=13,axis=1,hgap=1
)
row_ha.show_ticklabels(df.index.tolist(),fontdict={'color':'blue'},rotation=30)
plt.show()
# Here, we can use hgap (when axis=1) or wgap (when axis=0) to control the widh of height space between different annotations.
Starting plotting HeatmapAnnotations
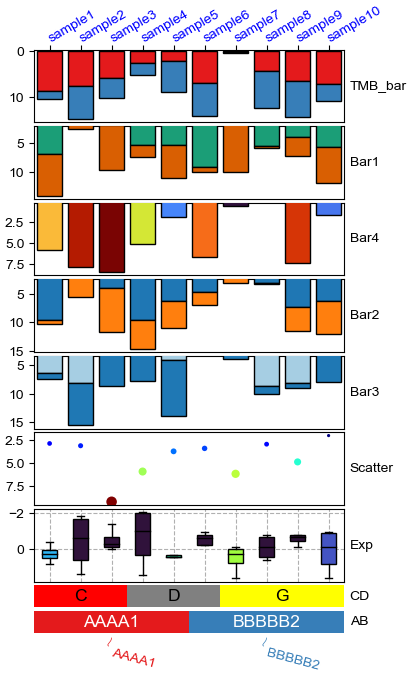
Change orentation to the left¶
[12]:
plt.figure(figsize=(8, 4))
row_ha = HeatmapAnnotation(
label=anno_label(df.AB, merge=True,rotation=15),
AB=anno_simple(df.AB,add_text=True,legend=True,
#text_kws=dict(bbox={"pad":0},va='center',ha='center',rotation_mode='anchor')
),
CD=anno_simple(df.CD,add_text=True,legend=True),
Exp=anno_boxplot(df_box, cmap='turbo',legend=True),
Scatter=anno_scatterplot(df_scatter),
TMB_bar=anno_barplot(df_bar,legend=True,cmap='Set1'),
Bar1=anno_barplot(df_bar1,legend=True,cmap='Dark2'),
Bar4=anno_barplot(df_bar4,legend=True,cmap='turbo'),
Bar2=anno_barplot(df_bar2,legend=True,cmap='tab10'),
Bar3=anno_barplot(df_bar3,legend=True,cmap='Paired'),
plot=True,legend=True,legend_gap=5,
axis=0,legend_hpad=20,label_side='bottom',wgap=3,
)
row_ha.show_ticklabels(df.index.tolist(),fontdict={'color':'blue'},rotation=0)
plt.show()
Starting plotting HeatmapAnnotations
Collecting annotation legends..
Incresing ncol
Incresing ncol
Incresing ncol
More than 3 cols is not supported
Legend too long, generating a new column..
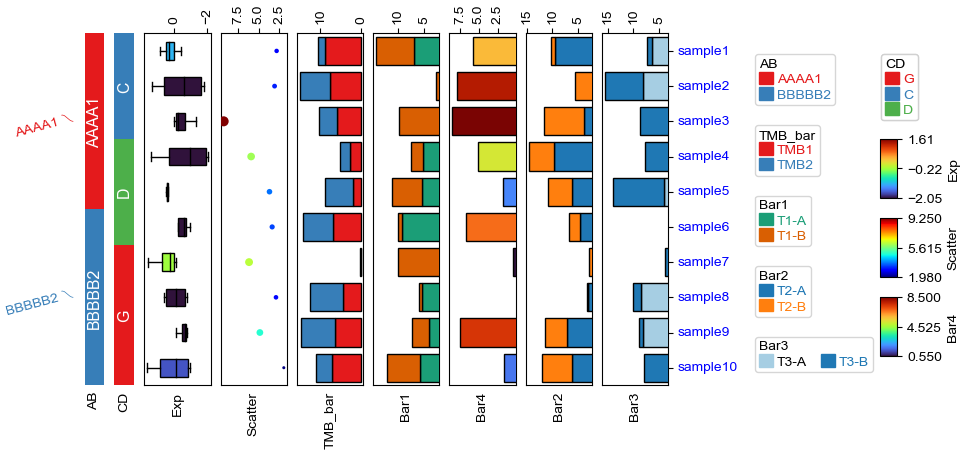
Change orentation to the right¶
[13]:
plt.figure(figsize=(8, 4))
row_ha = HeatmapAnnotation(
TMB_bar=anno_barplot(df_bar,legend=True,cmap='Set1'),
Bar1=anno_barplot(df_bar1,legend=True,cmap='Dark2'),
Bar4=anno_barplot(df_bar4,legend=True,cmap='turbo'),
Bar2=anno_barplot(df_bar2,legend=True,cmap='tab10'),
Bar3=anno_barplot(df_bar3,legend=True,cmap='Paired'),
Scatter=anno_scatterplot(df_scatter,grid=True),
Exp=anno_boxplot(df_box, cmap='turbo',legend=True,grid=True),
CD=anno_simple(df.CD, colors={'C': 'red', 'D': 'gray', 'G': 'green'},
add_text=True,legend=True,
text_kws={'rotation':-90}),
AB=anno_simple(df.AB,add_text=True,legend=True,
text_kws={'rotation':-90,'color':'black'}),
label=anno_label(df.AB, merge=True,rotation=15),
plot=True,legend=True,legend_hpad=13,legend_gap=5,axis=0,wgap=3,
)
row_ha.show_ticklabels(df.index.tolist(),fontdict={'color':'black'},rotation=0)
plt.show()
Starting plotting HeatmapAnnotations
Collecting annotation legends..
Incresing ncol
Incresing ncol
Incresing ncol
More than 3 cols is not supported
Legend too long, generating a new column..
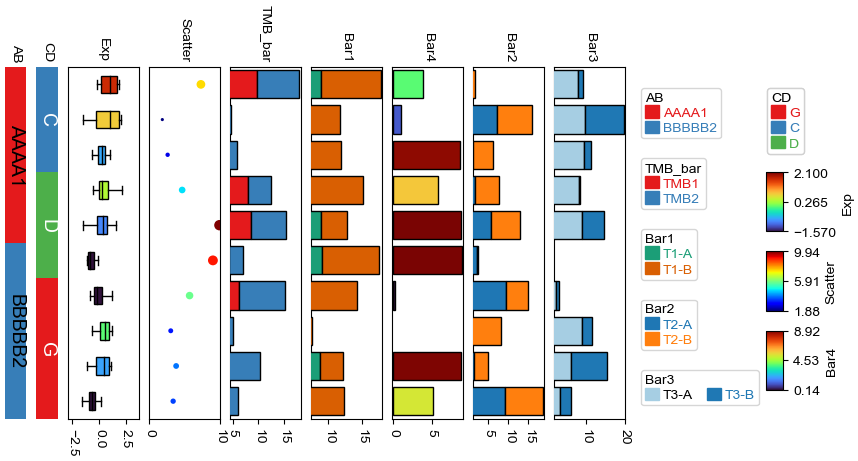
Changing orientation using parameter orientation¶
By Default, if there is no anno_label in the annotation, the oriention would be determined by parameter orientation.
[14]:
plt.figure(figsize=(8, 4))
col_ha = HeatmapAnnotation(
AB=anno_simple(df.AB,add_text=True,legend=True),
CD=anno_simple(df.CD,add_text=True,legend=True),
Exp=anno_boxplot(df_box, cmap='turbo',legend=True),
Scatter=anno_scatterplot(df_scatter,grid=True),
TMB_bar=anno_barplot(df_bar,legend=True,cmap='Set1'),
Bar1=anno_barplot(df_bar1,legend=True,cmap='Dark2'),
Bar4=anno_barplot(df_bar4,legend=True,cmap='turbo'),
Bar2=anno_barplot(df_bar2,legend=True,cmap='tab10'),
Bar3=anno_barplot(df_bar3,legend=True,cmap='Paired'),
plot=True,legend=True,axis=0,
legend_gap=5,orientation='left',
)
plt.show()
Starting plotting HeatmapAnnotations
Collecting annotation legends..
Incresing ncol
Incresing ncol
Incresing ncol
More than 3 cols is not supported
Legend too long, generating a new column..
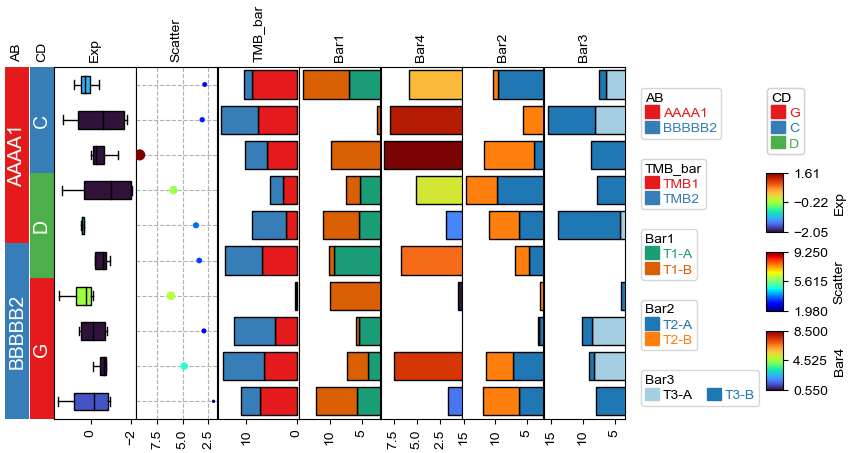
[15]:
plt.figure(figsize=(8, 4))
col_ha = HeatmapAnnotation(
AB=anno_simple(df.AB,add_text=True,legend=True,
text_kws={'rotation':-90,'fontsize':14,'color':'black'}),
CD=anno_simple(df.CD,add_text=True,legend=True,
text_kws={'rotation':-90,'fontsize':14,'color':'white'}),
Exp=anno_boxplot(df_box, cmap='turbo',legend=True),
Scatter=anno_scatterplot(df_scatter),
TMB_bar=anno_barplot(df_bar,legend=True,cmap='Set1'),
Bar1=anno_barplot(df_bar1,legend=True,cmap='Dark2'),
Bar4=anno_barplot(df_bar4,legend=True,cmap='turbo'),
Bar2=anno_barplot(df_bar2,legend=True,cmap='tab10'),
Bar3=anno_barplot(df_bar3,legend=True,cmap='Paired'),
plot=True,legend=True,axis=0,wgap=3,
legend_gap=5,orientation='right',
)
plt.show()
Starting plotting HeatmapAnnotations
Collecting annotation legends..
Incresing ncol
Incresing ncol
Incresing ncol
More than 3 cols is not supported
Legend too long, generating a new column..
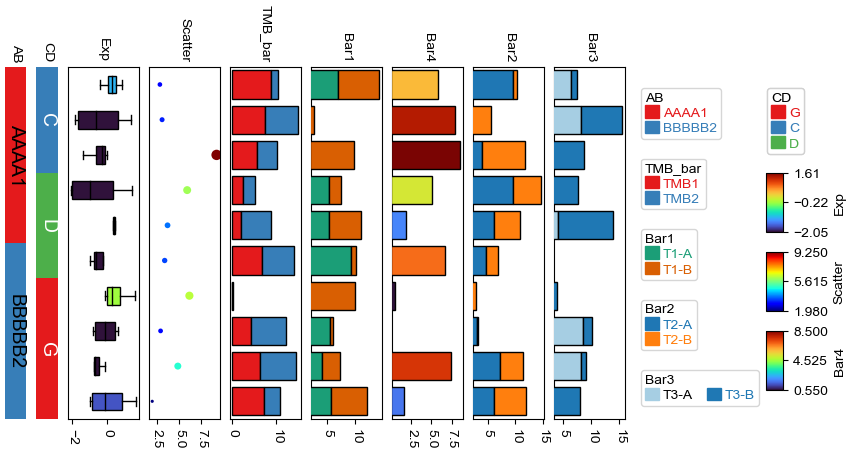
Add multiple heatmap annotations using for loop¶
Typically, we can create a heatmap annotatin using the following code:
col_ha = HeatmapAnnotation(
Group=anno_simple(df_cols.hypomethylated_samples,colors=sample_group_color_dict,legend=True),
CellType=anno_simple(df_cols.CellType,colors=ct_color_dict,legend=ct_legend),
M1=anno_simple(df_cols['M1'],cmap='jet',legend=lgd,vmax=1,vmin=0,legend_kws={'label':'M1'}),
verbose=0,label_side='right',label_kws={'horizontalalignment':'left'})
But what if we have many annotations, for example:
col_ha = HeatmapAnnotation(
Group=anno_simple(df_cols.hypomethylated_samples,colors=sample_group_color_dict,legend=True),
CellType=anno_simple(df_cols.CellType,colors=ct_color_dict,legend=ct_legend),
M1=anno_simple(df_cols['M1'],cmap='jet',legend=lgd,vmax=1,vmin=0,legend_kws={'label':'M1'}),
M2=anno_simple(df_cols['M2'],cmap='jet',legend=lgd,vmax=1,vmin=0,legend_kws={'label':'M2'}),
M3=anno_simple(df_cols['M3'],cmap='jet',legend=lgd,vmax=1,vmin=0,legend_kws={'label':'M3'}),
.....
verbose=0,label_side='right',label_kws={'horizontalalignment':'left'})
In this case, we can create an dict including the name and annotation as keys and values:
col_ha_dict={
'Group':anno_simple(df_cols.hypomethylated_samples,colors=sample_group_color_dict,legend=True),
'CellType':anno_simple(df_cols.CellType,colors=ct_color_dict,legend=ct_legend)
}
for col in sample_cols:
col_ha_dict[col]=anno_simple(df_cols[col],cmap='jet',legend=lgd,vmax=1,vmin=0,legend_kws={'label':col})
col_ha = HeatmapAnnotation(**col_ha_dict,
verbose=0,label_side='right',label_kws={'horizontalalignment':'left'})
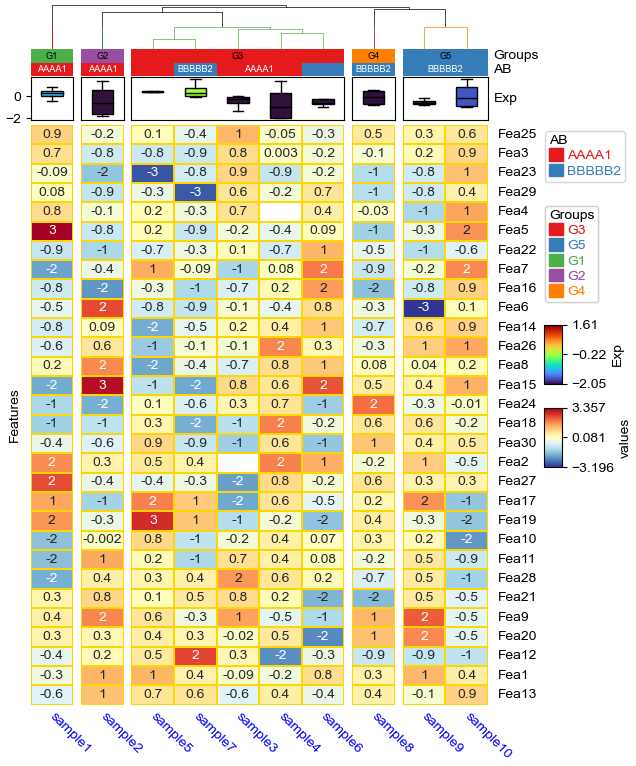
Cluster between groups and cluster within groups¶
Similar to cluster_between_groups and cluster_within_groups in R (https://jokergoo.github.io/2021/03/05/cluster-groups-in-complexheatmap/)
clsuter within groups: col_split=*, col_cluster=True¶
[16]:
df['Groups']=['G1']+['G2']+['G3']*5+['G4']+['G5']*2
col_ha = HeatmapAnnotation(
Groups=anno_simple(df.Groups,add_text=True,text_kws={'color':'black'}),
AB=anno_simple(df.AB,add_text=True),axis=1,
Exp=anno_boxplot(df_box, cmap='turbo'),
verbose=0) #verbose=0 will turn off the log.
plt.figure(figsize=(6, 8))
cm = ClusterMapPlotter(
data=df_heatmap, top_annotation=col_ha,
col_split=df.Groups,col_split_gap=2,
col_cluster=True,row_cluster=True,col_dendrogram=True,
label='values',show_rownames=True,show_colnames=True,
tree_kws={'col_cmap': 'Set1'},verbose=0,legend_gap=7,
annot=True,fmt='.1g',linewidths=0.05,linecolor='gold',cmap='RdYlBu_r',
xticklabels_kws={'labelrotation':-45,'labelcolor':'blue'},
ylabel='Features',
legend_order=['AB','Groups','Exp','values'] #change legend order
)
plt.show()
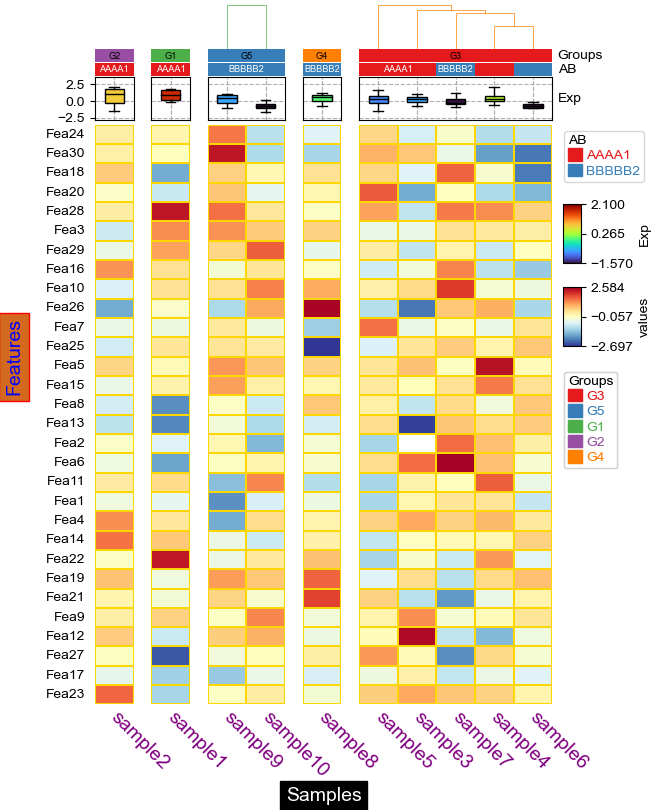
cluster_between_groups: col_split=*, col_split_order="cluster_between_groups",col_cluster=False¶
[17]:
plt.figure(figsize=(8, 10))
cm = ClusterMapPlotter(
data=df_heatmap, top_annotation=col_ha,
col_split=df.Groups, col_split_order="cluster_between_groups",
col_split_gap=2,col_cluster=False,
row_cluster=True,col_dendrogram=True,row_dendrogram_size=35,col_dendrogram_size=25,
row_split=2,row_split_gap=1,row_dendrogram=True,
label='values',show_rownames=True,show_colnames=True,bezier=True,dotsize=8,
tree_kws={'colors':'blue','row_cmap':'Set1','col_cmap':'Paired'},
verbose=0,legend_gap=7,
linewidths=0.05,linecolor='gold',cmap='RdYlBu_r',
xticklabels_kws={'labelrotation':-45,'labelcolor':'blue'},
ylabel='Features')
plt.show()
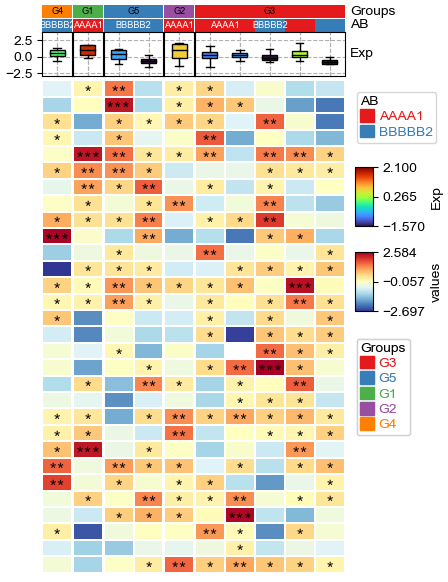
cluster_within_groups && cluster_between_groups: col_split=*, col_split_order="cluster_between_groups",col_cluster=True¶
[18]:
plt.figure(figsize=(6, 8))
cm = ClusterMapPlotter(
data=df_heatmap, top_annotation=col_ha,
col_split=df.loc[:,['AB','Groups']], col_split_order="cluster_between_groups",
col_split_gap=2,col_cluster=True,row_split_gap=1.5,
row_split=3,#row_split_order='cluster_between_groups',
row_cluster=True,col_dendrogram=True,row_dendrogram=True,
label='values',show_rownames=True,show_colnames=True,
tree_kws={'colors':'blue'},verbose=0,legend_gap=7,
annot=True,fmt='.1g',linewidths=0.05,linecolor='gold',cmap='RdYlBu_r',
xticklabels_kws={'labelrotation':-45,'labelcolor':'blue'},
ylabel='Features')
plt.show()
[19]:
# `label_kws` in `HeatmapAnnotation` control the heatmap annotaiton labels
col_ha = HeatmapAnnotation(
Groups=anno_simple(df.Groups,add_text=True,text_kws={'color':'black'}),
AB=anno_simple(df.AB,add_text=True),axis=1,
Exp=anno_boxplot(df_box, cmap='turbo',grid=True),
verbose=0,label_side='right'
)
# `xticklabels_kws` and `yticklabels_kws` control the ticklabels for the heatmap.
plt.figure(figsize=(6, 8))
cm = ClusterMapPlotter(data=df_heatmap, top_annotation=col_ha,
col_split=df.Groups,col_split_order=['G2','G1','G5','G4','G3'],
col_split_gap=4.5,col_cluster=True,
row_cluster=True,col_dendrogram=True,
label='values',show_rownames=True,show_colnames=True,
row_names_side='left',
tree_kws={'col_cmap':'Set1'},verbose=0,legend_gap=7,
linewidths=0.05,linecolor='gold',cmap='RdYlBu_r',
xticklabels_kws=dict(labelrotation=-45,labelcolor='purple',labelsize=14),
#more parameters for [x/y]_ticklabels_kws, see: matplotlib.axes.Axes.tick_params or ?ax.tick_params
xlabel='Samples',ylabel="Features",
xlabel_kws=dict(color='white',fontsize=14),
ylabel_kws=dict(color='blue',fontsize=14,labelpad=45), #increace labelpad manually using labelpad (points)
xlabel_bbox_kws=dict(facecolor='black'),
ylabel_bbox_kws=dict(facecolor='chocolate',edgecolor='red'),
)
plt.savefig("test.pdf",bbox_inches='tight')
plt.show()
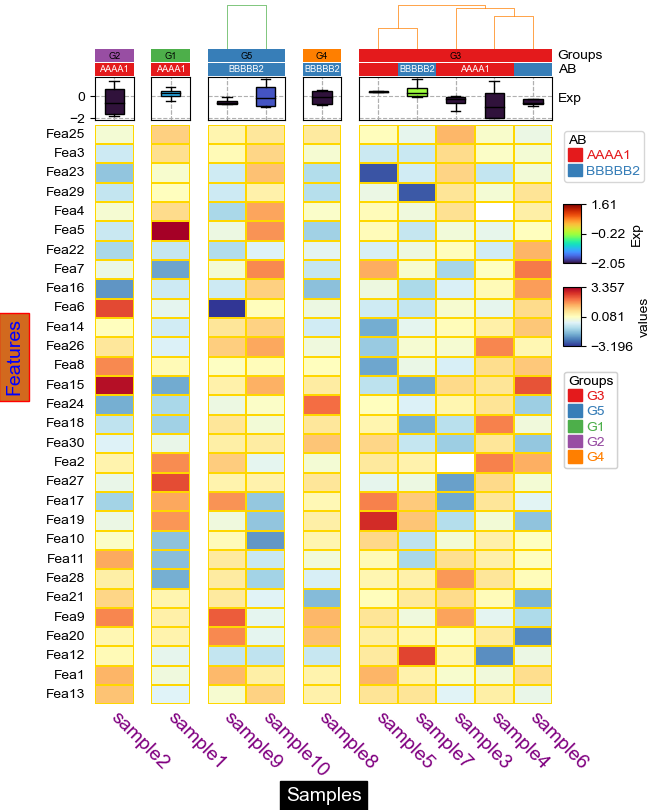
Custom annotation¶
[20]:
annot=df_heatmap.applymap(lambda x:'∗∗∗' if x >= 2 else '∗∗' if x >=1 else '∗' if x >0 else '')
# To make asterisk located at center in vertical, use ∗ ASTERISK OPERATOR. instead of normal *; see: https://unicode-explorer.com/c/2217
plt.figure(figsize=(5, 6.5))
cm = ClusterMapPlotter(
data=df_heatmap, top_annotation=col_ha,
annot=annot,fmt=None,annot_kws={'color':'white','fontname':'Courier'},
col_split=df.Groups, col_split_order="cluster_between_groups",
col_cluster=True,row_cluster=True,
label='values',
tree_kws={'col_cmap': 'Set1'},verbose=0,legend_gap=7,
linewidths=0.05,linecolor='white',cmap='RdYlBu_r',
xticklabels_kws={'labelrotation':-45,'labelcolor':'blue'})
plt.show()
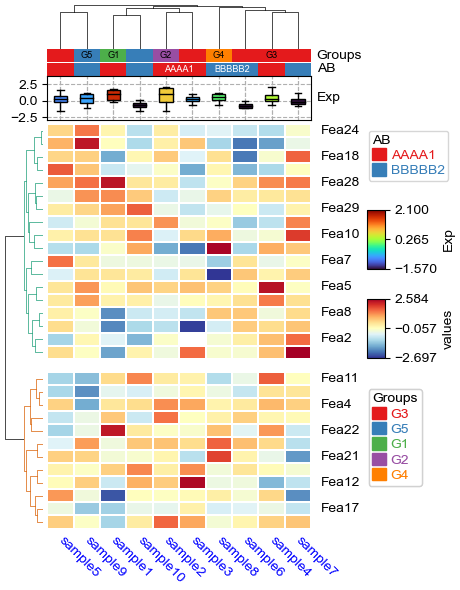
Custom linkage¶
[21]:
import fastcluster
# custom column linkage
linkage = fastcluster.linkage(df_heatmap.T.apply(lambda x:x.fillna(x.median()),axis=1), method='average', metric='canberra')
print("df_heatmap shape:",df_heatmap.shape,"\nlinkage shape:",linkage.shape,"\n",linkage)
plt.figure(figsize=(4, 6))
cm = ClusterMapPlotter(
data=df_heatmap, top_annotation=col_ha,z_score=0,
col_cluster=True,row_cluster=True,show_rownames=True,show_colnames=True,
label='values',col_dendrogram_kws=dict(linkage=linkage),col_dendrogram=True,
tree_kws={'col_cmap': 'Set1'},verbose=0,legend_gap=7,
linewidths=0.05,linecolor='white',cmap='RdYlBu_r',
xticklabels_kws={'labelrotation':-45,'labelcolor':'blue'})
plt.show()
df_heatmap shape: (30, 10)
linkage shape: (9, 4)
[[ 1. 6. 16.39057782 2. ]
[ 3. 8. 16.59636466 2. ]
[ 7. 11. 18.67771195 3. ]
[ 4. 10. 19.30874384 3. ]
[ 5. 9. 19.51326053 2. ]
[ 0. 2. 20.63068239 2. ]
[12. 13. 20.65079744 6. ]
[15. 16. 22.1959461 8. ]
[14. 17. 23.33605841 10. ]]
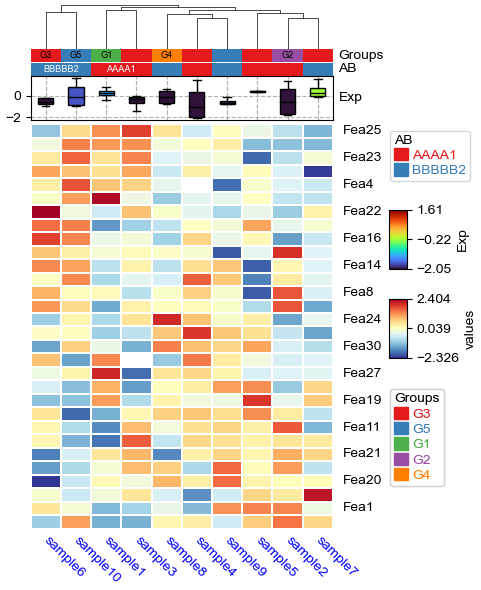
[22]:
df['Groups']=['G1']+['G2']+['G3']*5+['G4']+['G5']*2
plt.figure(figsize=(4, 6))
cm = ClusterMapPlotter(
data=df_heatmap, top_annotation=col_ha,
col_cluster=True,row_cluster=True,show_rownames=True,show_colnames=True,
row_split=2,row_split_gap=3,row_dendrogram=True,
label='values',col_dendrogram_kws=dict(linkage=linkage),col_dendrogram=True,
tree_kws={'col_cmap': 'Set1','row_cmap':'Dark2'},verbose=0,legend_gap=7,
linewidths=0.05,linecolor='white',cmap='RdYlBu_r',
xticklabels_kws={'labelrotation':-45,'labelcolor':'blue'})
plt.show()
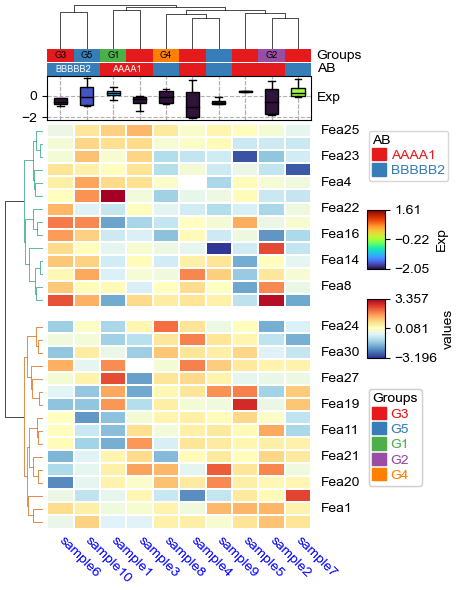
Image annotation¶
[23]:
df = pd.DataFrame(['AAAA1'] * 5 + ['BBBBB2'] * 5, columns=['AB'])
df['CD'] = ['C'] * 3 + ['D'] * 3 + ['G'] * 4
df['F'] = np.random.normal(0, 1, 10)
df.index = ['sample' + str(i) for i in range(1, df.shape[0] + 1)]
df_box = pd.DataFrame(np.random.randn(10, 4), columns=['Gene' + str(i) for i in range(1, 5)])
df_box.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar = pd.DataFrame(np.random.uniform(0, 10, (10, 2)), columns=['TMB1', 'TMB2'])
df_bar.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_scatter = pd.DataFrame(np.random.uniform(0, 10, 10), columns=['Scatter'])
df_scatter.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar1 = pd.DataFrame(np.random.uniform(0, 10, (10, 2)), columns=['T1-A', 'T1-B'])
df_bar1.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar2 = pd.DataFrame(np.random.uniform(0, 10, (10, 2)), columns=['T2-A', 'T2-B'])
df_bar2.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar3 = pd.DataFrame(np.random.uniform(0, 10, (10, 2)), columns=['T3-A', 'T3-B'])
df_bar3.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar3.iloc[5,0]=np.nan
df_bar4 = pd.DataFrame(np.random.uniform(0, 10, (10, 1)), columns=['T4'])
df_bar4.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar4.iloc[7,0]=np.nan
df_img = pd.DataFrame(['https://upload.wikimedia.org/wikipedia/commons/thumb/c/c3/Python-logo-notext.svg/121px-Python-logo-notext.svg.png',
'https://motifcollections.aertslab.org/v10nr_clust/logos/metacluster_135.7.png',
'https://cdn3.iconfinder.com/data/icons/family-member-flat-happy-family-day/512/Brother-512.png',
'https://cdn3.iconfinder.com/data/icons/family-member-flat-happy-family-day/512/Sister-512.png',
'https://img.freepik.com/free-vector/sticker-design-with-cute-mouse-isolated_1308-59360.jpg',
'https://motifcollections.aertslab.org/v10nr_clust/logos/metacluster_131.8.png',
'https://img.freepik.com/premium-vector/vector-illustration-gorilla-isolated-white-background-cartoon-style_1151-66575.jpg',
"2.png",'1.jpeg',
'https://upload.wikimedia.org/wikipedia/commons/thumb/c/c3/Python-logo-notext.svg/121px-Python-logo-notext.svg.png'], columns=['path'])
df_img.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
[24]:
plt.figure(figsize=(16, 4))
col_ha = HeatmapAnnotation(
label=anno_label(df.AB, merge=True,rotation=15),
AB=anno_simple(df.AB,add_text=True,legend=True), axis=1,
CD=anno_simple(df.CD, add_text=True,legend=True,text_kws={'color':'black'}),
Exp=anno_boxplot(df_box, cmap='turbo',legend=True),
Scatter=anno_scatterplot(df_scatter),
Bar1=anno_barplot(df_bar1,legend=True,cmap='Dark2'),
Bar4=anno_barplot(df_bar4,legend=True,cmap='turbo'),
Img=anno_img(df_img.path,border_width=5,border_color=255,height=15),
plot=True,legend=True,legend_gap=5,hgap=0.5)
col_ha.show_ticklabels(df.index.tolist(),fontdict={'color':'blue'},rotation=-30)
plt.show()
Starting plotting HeatmapAnnotations
Collecting annotation legends..
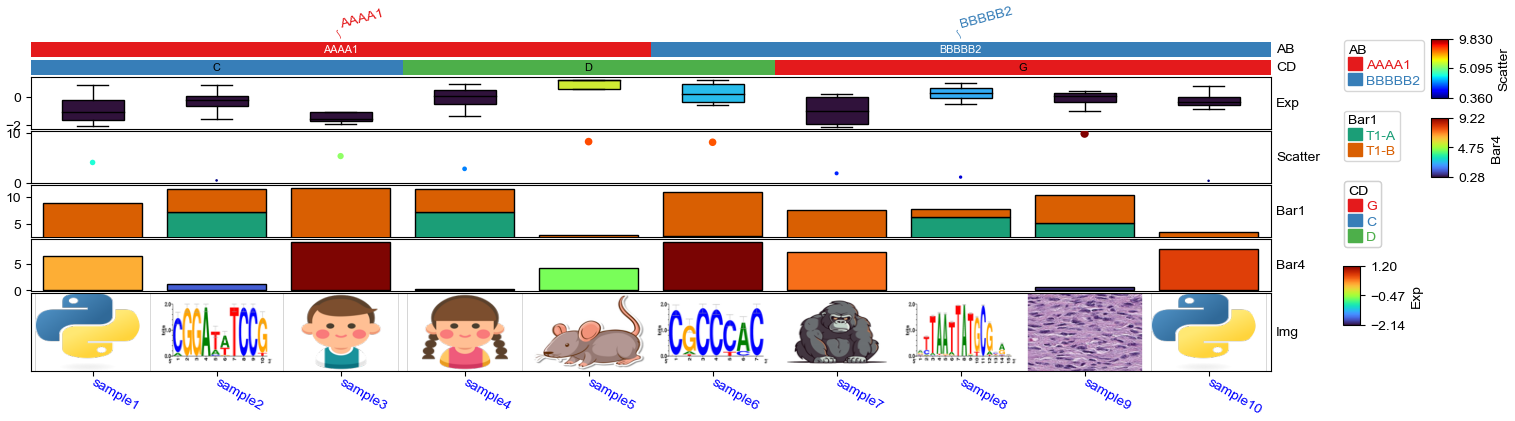
How to force display all row/col ticklabels?¶
When the height or width is not big enough to display all xticklabels and yticklabels, some ticklabels will be hidden to avoid overlapping. For example:
[25]:
plt.figure(figsize=(3.5, 5))
cm = ClusterMapPlotter(
data=df_heatmap,
col_cluster=True,row_cluster=True,
col_split=df.AB,row_split=2,
col_split_gap=0.5,row_split_gap=0.8,
label='values',row_dendrogram=True,
show_rownames=True,show_colnames=True,row_names_side='right',
tree_kws={'row_cmap': 'Set1'},verbose=0,legend_gap=5,
cmap='meth2',xticklabels_kws={'labelrotation':-90,'labelcolor':'blue'})
plt.show()
To force display all ticklabels no matter whether the height or width is big enough, set parameters xticklabels/yticklabels to True:
[26]:
plt.figure(figsize=(3.5, 5))
cm = ClusterMapPlotter(
data=df_heatmap,
col_cluster=True,row_cluster=True,
col_split=df.AB,row_split=2,
col_split_gap=0.5,row_split_gap=0.8,
label='values',row_dendrogram=True,
show_rownames=True,show_colnames=True,
row_names_side='right',yticklabels=True,
tree_kws={'row_cmap': 'Set1'},verbose=0,legend_gap=5,
cmap='meth2',xticklabels_kws={'labelrotation':-90,'labelcolor':'blue'})
plt.show()