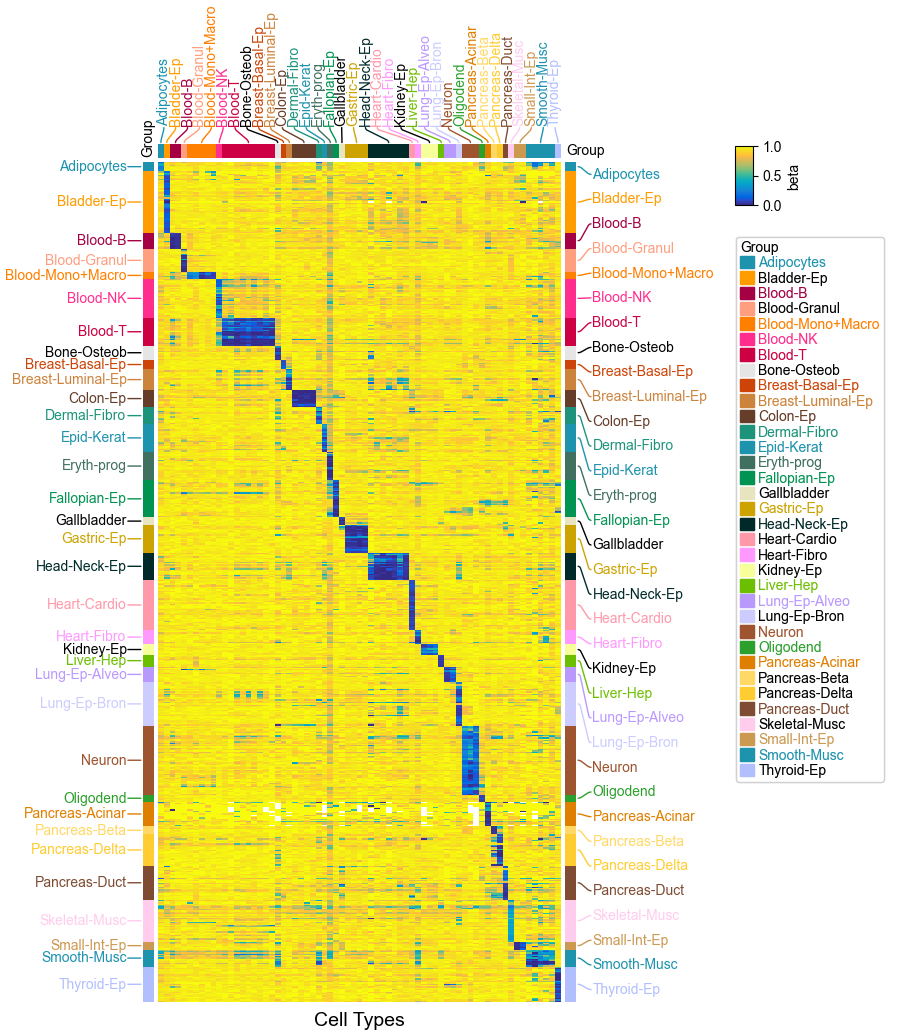
Visualizing Single Cell DNA Methylation Data (Loyfer2023)¶
[1]:
import os,sys
%matplotlib inline
import matplotlib.pylab as plt
import pickle
plt.rcParams['figure.dpi'] = 100
plt.rcParams['savefig.dpi']=300
plt.rcParams['font.family']='sans serif'
plt.rcParams['font.sans-serif']='Arial'
plt.rcParams['pdf.fonttype']=42
sys.path.append(os.path.expanduser("~/Projects/Github/PyComplexHeatmap/"))
import PyComplexHeatmap as pch
pch.use_pch_style() # or plt.style.use('default') to restore default style
print(pch.__version__)
1.7.8.dev6+gbc4db3a.d20241016
Read Loyfer2023 DNA methylation dataset (cell types level for cell type methylation signatures) Loyfer2023 DNA methylation dataset comes from PMID: 36599988
[2]:
data=pd.read_csv("https://raw.githubusercontent.com/DingWB/PyComplexHeatmap/main/data/Loyfer2023.meth.csv",sep='\t',index_col=0)
df_row=pd.read_csv("https://raw.githubusercontent.com/DingWB/PyComplexHeatmap/main/data/Loyfer2023.meth.rows.csv",sep='\t',index_col=0)
df_col=pd.read_csv("https://raw.githubusercontent.com/DingWB/PyComplexHeatmap/main/data/Loyfer2023.meth.cols.csv",sep='\t',index_col=0)
[3]:
data.head()
[3]:
| Adipocytes | Bladder-Epithelium | Blood-B-Mem | Blood-B | Blood-Granulocytes | Colon-Macrophages | Liver-Macrophages | Blood-Monocytes | Lung-Interstitial-Macrophages | Lung-Alveolar-Macrophages | ... | Pancreas-Duct | Skeletal-Muscle | Small-int-Epithelium | Small-int-Endocrine | Lung-Bronchus-Smooth-Muscle | Prostate-Smooth-Muscle | Bladder-Smooth-Muscle | Coronary-Artery-Smooth-Muscle | Aorta-Smooth-Muscle | Thyroid-Epithelium | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr1-110426397 | 0.120667 | 0.9102 | 0.8285 | 0.918667 | 0.750667 | 0.8705 | 0.896 | 0.916333 | 0.953333 | 0.9450 | ... | 0.61225 | 0.5485 | 0.94175 | 0.952 | 0.417 | 0.150 | 0.229 | 0.294 | 0.214 | 0.938333 |
| chr1-110426639 | 0.420000 | 0.9304 | 0.8920 | 0.955667 | 0.964667 | 0.9285 | 0.903 | 0.924667 | 0.975333 | 0.9720 | ... | 0.94575 | 0.8990 | 0.96225 | 0.963 | 0.606 | 0.185 | 0.348 | 0.917 | 0.484 | 0.972333 |
| chr1-110426659 | 0.425667 | 0.9328 | 0.7635 | 0.933000 | 0.942333 | 0.8945 | 0.931 | 0.963667 | 0.899333 | 0.8895 | ... | 0.93875 | 0.8390 | 0.92225 | 0.903 | 0.559 | 0.129 | 0.292 | 0.875 | 0.395 | 0.989667 |
| chr1-110427109 | 0.203667 | 0.9724 | 0.9610 | 0.963667 | 0.968667 | 0.9240 | 0.939 | 0.985000 | 0.968667 | 0.9620 | ... | 0.73300 | 0.7570 | 0.96525 | 0.939 | 0.929 | 0.225 | 0.806 | 0.951 | 0.905 | 0.974333 |
| chr1-110427116 | 0.217333 | 0.9478 | 1.0000 | 0.976000 | 0.970667 | 0.9730 | 0.913 | 0.975000 | 0.970333 | 0.9800 | ... | 0.65125 | 0.7955 | 0.96150 | 1.000 | 0.952 | 0.605 | 0.857 | 1.000 | 0.868 | 0.981000 |
5 rows × 69 columns
[4]:
df_row.head()
[4]:
| Group | |
|---|---|
| ID | |
| chr1-110426397 | Adipocytes |
| chr1-110426639 | Adipocytes |
| chr1-110426659 | Adipocytes |
| chr1-110427109 | Adipocytes |
| chr1-110427116 | Adipocytes |
[5]:
df_col.head()
[5]:
| Group | |
|---|---|
| ClusterID | |
| Adipocytes | Adipocytes |
| Bladder-Epithelium | Bladder-Ep |
| Blood-B-Mem | Blood-B |
| Blood-B | Blood-B |
| Blood-Granulocytes | Blood-Granul |
[6]:
df_col.Group.unique()
[6]:
array(['Adipocytes', 'Bladder-Ep', 'Blood-B', 'Blood-Granul',
'Blood-Mono+Macro', 'Blood-NK', 'Blood-T', 'Bone-Osteob',
'Breast-Basal-Ep', 'Breast-Luminal-Ep', 'Colon-Ep', 'Dermal-Fibro',
'Epid-Kerat', 'Eryth-prog', 'Fallopian-Ep', 'Gallbladder',
'Gastric-Ep', 'Head-Neck-Ep', 'Heart-Cardio', 'Heart-Fibro',
'Kidney-Ep', 'Liver-Hep', 'Lung-Ep-Alveo', 'Lung-Ep-Bron',
'Neuron', 'Oligodend', 'Pancreas-Acinar', 'Pancreas-Beta',
'Pancreas-Delta', 'Pancreas-Duct', 'Skeletal-Musc', 'Small-Int-Ep',
'Smooth-Musc', 'Thyroid-Ep'], dtype=object)
[7]:
col_colors_dict={
'Adipocytes':'#1E93AE','Bladder-Ep':'#FF9C00','Blood-B':'#A40043','Blood-Granul':'#FF9F7F',
'Blood-Mono+Macro':'#FF7F00','Blood-NK':'#FF2E8D','Blood-T':'#CC0043','Bone-Osteob':'#E5E5E5',
'Breast-Basal-Ep':'#CC4407','Breast-Luminal-Ep':'#CC843D','Colon-Ep':'#663D28','Dermal-Fibro':'#1E937C',
'Epid-Kerat':'#1E93AE','Eryth-prog':'#40705F','Fallopian-Ep':'#009351','Gallbladder':'#E7E4BF',
'Gastric-Ep':'#CCA300','Head-Neck-Ep':'#002929','Heart-Cardio':'#FF99AA','Heart-Fibro':'#FF99FF',
'Kidney-Ep':'#F6FF99','Liver-Hep':'#6CBF00','Lung-Ep-Alveo':'#BA99FF','Lung-Ep-Bron':'#CCCCFF',
'Neuron':'#9e542e','Oligodend':'#2ca02c','Pancreas-Acinar':'#DF7F00','Pancreas-Beta':'#FFD866',
'Pancreas-Delta':'#FFCC32','Pancreas-Duct':'#7F4C33','Skeletal-Musc':'#FFCCEE','Small-Int-Ep':'#CC9951',
'Smooth-Musc':'#1E93AE','Thyroid-Ep':'#B2BFFF'}
col_ha = pch.HeatmapAnnotation(label=pch.anno_label(df_col['Group'],merge=True,rotation=90,extend=True,
colors=col_colors_dict,adjust_color=True,luminance=0.75,
relpos=(0.5,0)), #fontsize=10
Group=pch.anno_simple(df_col['Group'],colors=col_colors_dict), #legend_kws={'fontsize':4}
verbose=1,axis=1)
left_ha = pch.HeatmapAnnotation(
label=pch.anno_label(df_row['Group'],merge=True,extend=False,
colors=col_ha.annotations[1].color_dict,adjust_color=True,luminance=0.75,
relpos=(1,0.5)),
Group=pch.anno_simple(df_row['Group'],legend=True,
colors=col_ha.annotations[1].color_dict),
verbose=1,axis=0,plot_legend=False)
right_ha = pch.HeatmapAnnotation(
Group=pch.anno_simple(df_row['Group'],legend=True,
colors=col_ha.annotations[1].color_dict),
label=pch.anno_label(df_row['Group'],merge=True,extend=True,
colors=col_ha.annotations[1].color_dict,adjust_color=True,luminance=0.75,
relpos=(0,0.5)), #fontsize=10
verbose=1,axis=0,plot_legend=False,label_kws=dict(visible=False))
#data was sorted to have the same Group order for rows and columns
# df_row.sort_values('Group',inplace=True)
# df_col.sort_values('Group',inplace=True)
plt.figure(figsize=(6, 10))
cm = pch.ClusterMapPlotter(data=data.loc[df_row.index.tolist(),df_col.index.tolist()],
top_annotation=col_ha, left_annotation=left_ha,
right_annotation=right_ha,
row_cluster=False,col_cluster=False,
label='beta', row_dendrogram=False,legend_gap=7,
# row_split=df_row.Group,col_split=df_col.Group,
# row_split_gap=0.2,col_split_gap=0.1
# row_split_order=df_row.Group.unique().tolist(),
# col_split_order=df_row.Group.unique().tolist(),
cmap='parula',rasterized=True,
xlabel="Cell Types", legend_hpad=0,
# ylabel="CpGs",
xlabel_kws=dict(color='black', fontsize=14, labelpad=0),
# ylabel_kws=dict(color='black', fontsize=14, labelpad=0),
)
plt.savefig("Loyfer2023_heatmap.pdf",bbox_inches='tight')
plt.show()
Starting plotting..
Starting calculating row orders..
Reordering rows..
Starting calculating col orders..
Reordering cols..
Plotting matrix..
Starting plotting HeatmapAnnotations
Starting plotting HeatmapAnnotations
Starting plotting HeatmapAnnotations
Collecting legends..
Collecting annotation legends..
Collecting annotation legends..
Collecting annotation legends..
Plotting legends..
Estimated legend width: 41.981944444444444 mm